QualityControl4Genes
Even gene expression needs quality control!
With colleagues in Basel, we are looking further into some molecular mysteries around mRNA surveillance...
Interested to learn more?
Here is the link to the recent paper about one of the the human orthologues of this nématode gene, smg-4, and how it, hUPF3p-X is regulated by microRNAs and active in the brain. Regulation of genes in thymus and brain via the interplay of microRNA action on RNA surveillance factors affecting target gene expression was already shown by 2011.
As Nietzsche said - you have made your way from worm to man, but much within you is still worm...
This key pathway has the responsibility of removing mRNAs that might express dominant negative or toxic fragments of otherwise good genes. Errors are normal, and all the stages of gene expression - from transcription, to splicing, to translating to protein or activating other functions - can be subject to mistakes. Prevention is what mRNA surveillance is about. However, these genes also have developmental roles and natural targets which they regulate. More than 10% of the RNAs made in higher eukaryotes are affected by this system, and further gene expression regulation is possible even out in sub-cellular compartments, like dendrites...
There is even recent article in Trends in Neurosci about it all...
Here is the original paper about the cloned worm gene (Aronoff et al, 2001), smg-4, which defines a family of conserved genes.
In humans, there are two copies of this gene on different chromosomes, with the X-linked one tied to Intellectual Disabilities, and more interesting features in knockdown and attempts to knock this QC mechanism out in cells...
Additional feedback loops can affect activity or even expression levels, for appropriate function.
Here is part of an open access paper's introduction more focused on another quality control mechanism - but it all starts with mRNA surveillance (which from yeast studies had been also termed 'nonsense-mediated decay'):
Nonsense-mediated decay (NMD) (reviewed in [He and Jacobson, 2015]) is a translational surveillance pathway to mitigate deleterious products of premature stop codons. In NMD, recognition of an early stop codon destabilizes an mRNA (Morse and Yanofsky, 1969; Baserga and Benz, 1988; Losson and Lacroute, 1979). Foundational studies in S. cerevisiae and C. elegans revealed protein factors responsible for NMD (Leeds et al., 1991; Hodgkin et al., 1989; Pulak and Anderson, 1993). In the decades since, a large body of literature has highlighted similarities and differences in NMD between yeast and metazoans. For example, while both yeast and metazoan NMD involve a core set of three proteins (UPF1-3 in yeast, SMG-2–4 in metazoans), metazoans require additional proteins for NMD (e.g. SMG-1, –5, and −6). Additionally, Saccharomyces cerevisiae NMD is thought to occur predominantly through decapping and 5’>3’ exonucleolytic degradation (Muhlrad and Parker, 1994), while studies across metazoans have implicated both exo- and endonucleolytic machineries (e.g. [Lykke-Andersen, 2002; Lejeune et al., 2003; Gatfield and Izaurralde, 2004; Glavan et al., 2006; Huntzinger et al., 2008; Eberle et al., 2009; Lykke-Andersen et al., 2014; Schmidt et al., 2015; Ottens et al., 2017]).
Although protective under many circumstances, the NMD pathway also contributes to pathological suppression of expression from numerous disease-causing mutations (about 11% of point mutations responsible for human disease [Mort et al., 2008]).
(Selected References from the above paper excerpt, at bottom...)
Have questions?
Would you be interested to try some worm research?
To make it simple, genes like smg-4 (part of a genetic pathway conserved from yeast to humans) look for and eliminate aberrant messenger RNA transcripts with early 'stop' signals for the protein. Mistakes happen. Preventing expression of truncated proteins from such RNAs is however just part of what these genes do. As mentioned above, these systems also have 'natural' -often developmental- RNA targets.
Much more can be learned from the worm gene, and as part of this project, some progress has already been made.
Not only a Crispr knock-out and knock-in (of an mCherry tag into the gene), but also RNAi to 'knockdown' gene expression were planned for this molecular genetic project in the model system, C. elegans, in collaboration with the Mango lab at the Biozentrum of the University of Basel. While getting to plasmid clones from a yeast artificial chromosome was key to publish the smg-4 cloning paper, moving forward to using the latest tools in molecular biology (from the RNA world!) has been a great plus of this project.
Synthetic biology is amazing...
Injecting premade RNPs with guide RNAs and the repair template to make the KO was super efficient! (Of 25 initial transformants, 5 gave the perfect edit.)
The KO was already made by March 2023 !
We have already confirmed old results here, with 'rescue' by PCR products, as shown in the image above right...
(To note: 'rescue' makes the worms paralysed in this genetic context. It is just due to the fact that a myosin heavy chain gene is the target... We project a simpler explanation of it all, soon...)
The study began with rescue attempts using DNA fragments made by PCR, and attempts to KnockDown the gene with RNA interference (RNAi). Although the amplified transgenes worked a charm, products derived from mutant strains did not rescue, and had the sequences predicted, of course. For work published so long ago, we decided that finally the new tools of synthetic biology would being used. =)
Crispr based KnockOuts and a Knock-In have already been generated. However, in fact, the RNAi feeding, even in a super-sensitive mutant strain, rrf-3, did not give any effect.***
We also confirmed the deletion sequence of a mutant made with psoralen-uv, and this is interesting as it keeps the key domain, but the what is left of the gene only partially rescues (egg laying, but not muscle function).
The Knockout animals have Pvuls and act as a simple loss of function, so we hypothesize that at least one of the other genes in the operon have some essential function that makes it difficult to make new mutations in the region. One strain carrying the allele with a complex rearrangement is less fit than others, which also lends credence to this hypothesis. This sequence should be obtained, as possible. More molecular details can be seen here, for the smg-4 gene which forms the basis of the current C. elegans project #QualityControl4Genes in the Hackuarium P1.
Sequence confirmation on an old deletion in the gene (but much after the original paper) gives us clues about what is most important for function. We speculated some essential function, partly because mutations in the gene were rare (and the only point mutations found were on either end of the same intron...). A partial loss of function from a deletion made in Japan was also confirmed during this recent work.
Update March 2024
As mentioned above, the Knockout was successful on the first attempt, with 5 independent lines obtained from 25 initial 'roller' transformants. Crispr is amazing!
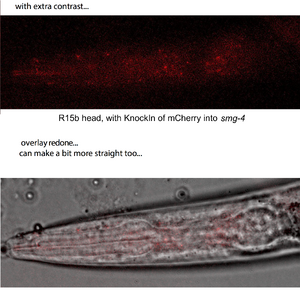
The KnockIn (to put the red mCherry marker, into the similar region as GFP was put in, for the original work 2 decades ago) was more of a challenge. Finally, a perfect insertion strain was obtained, and results with the KnockIn are just starting to come in (march24). Speckles in heads of animals are the key feature seen to date(more below). Outcrosses of the strain and crosses to put it in the context of a smg-suppressible myosin heavy chain mutation, unc-54(r293) have begun. In previous GFP fusions, the presence of target molecules apparently up regulate the expression, so muscle cells are expected to be seen in embryos again... Let's see!
Here is one of the first epi-fluor microscopy results (thanks to our friends at the UNIL):
speckles of the Smg-4 shuttling protein in a larval brain!
More experiments to come and writing of an article! Here is a recent image from the last trip up to Basel...
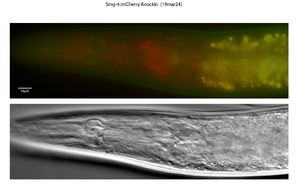
Hoping to learn much more in combination with other GFP tagged components of the pathway and nuclear (nuclear pore?) markers. If you would like to learn more too, or have ideas - do not hesitate to write! =)
Please contact rachel (at) hackuarium (dot) ch...
=)
***another gene mutant, SID-1, could still be tried for the RNAi feeding experiment given the results seen here, and the human UPF3-X function in the brain.
Nonetheless, with the KO a simple loss of function, and this KnockIn still to confirm for rescue of the unc-54(r293) allele, we can't help thinking how much we have to learn, still. How we can do all the experiments necessary, from RNA analyses to imaging (of subnuclear architecture and pore complexes and out to some potential outer nuclear membre cavity and ultimately the polyribosomes) in combination with green-tagged markers, will be a great challenge.
Maybe other collaborations could be possible - hoping that many labs will be interested...
Some References from the excerpt above:
He and Jacobson, 2015: Nonsense-Mediated mRNA Decay: Degradation of Defective Transcripts Is Only Part of the Story. https://doi.org/10.1146/annurev-genet-112414-054639
Morse and Yanofsky, 1969: Polarity and the Degradation of mRNA. https://doi.org/10.1038/224329a0
Baserga and Benz, 1988: Nonsense mutations in the human beta-globin gene affect mRNA metabolism. https://doi.org/10.1073/pnas.85.7.2056
Losson and Lacroute, 1979: Interference of nonsense mutations with eukaryotic messenger RNA stability. https://doi.org/10.1073/pnas.76.10.5134
Leeds et al, 1991: The product of the yeast UPF1 gene is required for rapid turnover of mRNAs containing a premature translational termination codon. doi:
10.1101/gad.5.12a.2303
Hodgkin et al, 1989: A new kind of informational suppression in the nematode Caenorhabditis elegans. doi: 10.1093/genetics/123.2.301
..:
Mort et al, 2008: A meta-analysis of nonsense mutations causing human genetic disease. https://doi.org/10.1002/humu.20763
Calixto et al 2010: Enhanced neuronal RNAi in C. elegans using SID-1.
https://doi.org/10.1038/nmeth.1463