Difference between revisions of "Automatic Cell Counter Algorithm"
| (12 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
| − | The aim of this algorithm is first to detect cells (or groups of cells), to count them and to find cells containing a micronucleus. Theoretically this | + | The aim of this algorithm is first to detect cells (or groups of cells), to count them and to find cells containing a micronucleus. Theoretically this may be easy as the micronuclei should be as coloured as the nucleus. But other elements, bacteria for instance, can also be coloured by the dye. The big deal seems to be able to distinguish micronuclei from those other elements. |
==Background and Material Used== | ==Background and Material Used== | ||
| Line 9: | Line 9: | ||
==Picture Procedure == | ==Picture Procedure == | ||
| − | First, we took photos while zooming on the cells of interest. As cells are quite translucent, | + | First, we took photos while zooming on the cells of interest. As cells are quite translucent, they were difficult to detect. In addition, we realized that the microscope was so dirty that the durst was detected as well as the cells and it may be difficult to distinguish them. It was thus decided to apply some pre-processing instead of directly searching for cells contour. |
| − | In order to apply the background removal, we took a picture of the background without focusing on anything. | + | The pre-processing will mainly remove noise due to the difference in luminosity. In addition it will ignore the durst that is present in both images. In order to apply the background removal, we took a picture of the background without focusing on anything and compare it to the image of interest. The pre-processing is easier if the field of view is the same on all images (same luminosity). Thus we thought it was better to take a picture of the whole field of view instead of zooming on the thing of interest. |
<gallery perrow="4" mode="packed" widths="px" heights="px"> | <gallery perrow="4" mode="packed" widths="px" heights="px"> | ||
| Line 20: | Line 20: | ||
==Cell Detection== | ==Cell Detection== | ||
| − | + | ===Pre-processing=== | |
| − | Another method was then tested, | + | Several methods were tried to remove the background from the image of interest. First a simple subtraction of the background image was done. Unfortunately, this method results in strange horizontals lines which may be due to the actualisation of the camera. For some images it was not a problem but for others the cell was very difficult to detect due to those lines. |
| + | Another method was then tested, the difference between the image representing the cell and the background image was computed. Then the cell is much more easily selected. | ||
<gallery perrow="4" mode="packed" widths="px" heights="px"> | <gallery perrow="4" mode="packed" widths="px" heights="px"> | ||
| Line 28: | Line 29: | ||
File:Difference.png|Difference Method | File:Difference.png|Difference Method | ||
</gallery> | </gallery> | ||
| + | |||
| + | ===Cell Selection=== | ||
Cell selection is done as followed: | Cell selection is done as followed: | ||
| Line 34: | Line 37: | ||
* Finally, the nucleus detection (next step) will be done on those larger contours only. | * Finally, the nucleus detection (next step) will be done on those larger contours only. | ||
| − | Results | + | ===Results=== |
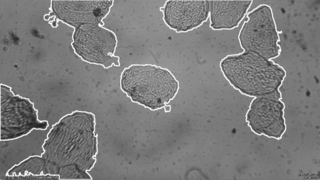
The developed algorithm is able to find isolated cells as well as groups of cells. Then a mask is applied in order to keep only what is inside the cells. This would simplify the next step which is finding the nucleus. | The developed algorithm is able to find isolated cells as well as groups of cells. Then a mask is applied in order to keep only what is inside the cells. This would simplify the next step which is finding the nucleus. | ||
<gallery perrow="4" mode="packed" widths="px" heights="px"> | <gallery perrow="4" mode="packed" widths="px" heights="px"> | ||
File:Screen Shot 2018-08-09 at 10.18.07.png|Single Cell Detection | File:Screen Shot 2018-08-09 at 10.18.07.png|Single Cell Detection | ||
| − | File: | + | File:Screen Shot 2018-09-03 at 17.19.26.png|Groups of Cells Detection |
</gallery> | </gallery> | ||
==Nucleus Detection== | ==Nucleus Detection== | ||
| + | |||
| + | ==Available Code== | ||
Latest revision as of 07:46, 4 September 2018
The aim of this algorithm is first to detect cells (or groups of cells), to count them and to find cells containing a micronucleus. Theoretically this may be easy as the micronuclei should be as coloured as the nucleus. But other elements, bacteria for instance, can also be coloured by the dye. The big deal seems to be able to distinguish micronuclei from those other elements.
Background and Material Used
Methylene blue is known as a DNA dye. Thus it enable to discriminate the nucleus from the rest of the cell. This dye is widely used and allow a quite good visual recognition. Micronuclei, which is also composed of DNA, will also be coloured by this dye.
Pictures used to build the algorithm are taken using the Raspberry Pi Camera connected to the microscope.
Picture Procedure
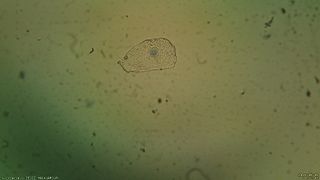
First, we took photos while zooming on the cells of interest. As cells are quite translucent, they were difficult to detect. In addition, we realized that the microscope was so dirty that the durst was detected as well as the cells and it may be difficult to distinguish them. It was thus decided to apply some pre-processing instead of directly searching for cells contour.
The pre-processing will mainly remove noise due to the difference in luminosity. In addition it will ignore the durst that is present in both images. In order to apply the background removal, we took a picture of the background without focusing on anything and compare it to the image of interest. The pre-processing is easier if the field of view is the same on all images (same luminosity). Thus we thought it was better to take a picture of the whole field of view instead of zooming on the thing of interest.
Cell Detection
Pre-processing
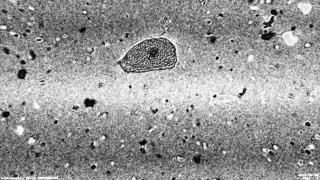
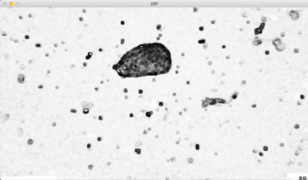
Several methods were tried to remove the background from the image of interest. First a simple subtraction of the background image was done. Unfortunately, this method results in strange horizontals lines which may be due to the actualisation of the camera. For some images it was not a problem but for others the cell was very difficult to detect due to those lines. Another method was then tested, the difference between the image representing the cell and the background image was computed. Then the cell is much more easily selected.
Cell Selection
Cell selection is done as followed:
- First contours on the difference image are computed.
- Then a thresholding step is done in order to keep only larger contours (larger area), the ones that may represent cells.
- Finally, the nucleus detection (next step) will be done on those larger contours only.
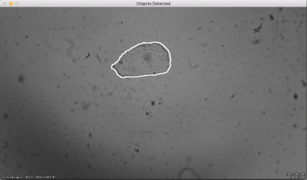
Results
The developed algorithm is able to find isolated cells as well as groups of cells. Then a mask is applied in order to keep only what is inside the cells. This would simplify the next step which is finding the nucleus.